Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Kim, O. T. P.*; Yura, Kei; Go, Nobuhiro
no journal, ,
Protein-RNA interactions play essential roles in a number of regulatory mechanisms of gene expression such as RNA splicing, transport and translation. Bioinformatics approach has been effective and rewarding in studying protein-nucleic acid interactions, especially protein-DNA. As the number of available protein-RNA complexes has increased, it is now possible to statistically examine protein-RNA interactions based on 3D structures. We carried out computational analyses of 86 non-homologous protein-RNA complexes retrieved fromP rotein Data Bank. Interface residue propensities, which give a measure forthe relative importance of different amino acid residues in the RNA-binding site, were calculated for both a single residue (singlet) and a residue pair(doublet). The result of singlet residue propensity showed that Arg, Lys, His, Phe, Tyr, Met and Gly were favored in RNA-binding site. The residue doublet interface propensity denoted the significant additional information compared with the sum of singlet propensities of the residues at protein-RNA interface. The interface residue propensities were used as parameters to predict protein-RNA interface. The accuracy of the prediction method was tested byjack-knife method, which attained more than 60% on average. The prediction method was applied to RNA-binding proteins with known 3D structures: mRNA export factors TAP, Mex67 and Mtr2. The prediction enables us to point out the candidate interfaces, which are consistent with previous studies and can betested by mutation experiments leading to elucidation of protein-RNA complexes.
Shionyu, Masafumi*; Yura, Kei; Hijikata, Atsushi*; Nakahara, Taku*; Shinoda, Kazuki*; Yamaguchi, Akihiro*; Takahashi, Kenichi*; Go, Michiko*
no journal, ,
Alternative splicing (AS) is a cellular process where multiple mature mRNAs are produced from a single gene by different usage of exons. From computational and experimental approaches, it is estimated that 30-70% of human genes undergo alternative splicing. There are a number of reports on spliced mRNAs involved in biological processes, yet functional analyses of a large number of proteins produced by AS remain to be performed. Functional analyses of these proteins by experiments are time-consuming, and therefore, a computational approach that can estimate effect of AS on proteins is required. We have developed a pipeline that can systematically detect AS regions, which are defined as protein regions modified by alternatively spliced exon, using genomic sequences and full-length transcripts data. The pipeline further assigned AS regions to protein three-dimensional structures and can estimate effects of AS on protein conformation stability and functional sites. We analyzed human AS regions using our pipeline and found that about half of AS regions fell into a size shorter than 100 amino acid residues. We, then, assessed the relationship between AS regions and protein structural domains and found that about 40% of AS regions were placed within a domain and the ratio of AS regions corresponding to domains was only about 10%. This result suggests that AS regulates protein function through alteration of segments within a domain rather than through switching protein domains. We will discuss how AS regulates protein function through alteration of segments within a domain.
 ; A Novel key protein "PprM"
; A Novel key protein "PprM"Oba, Hirofumi; Sghaier, H.; Sato, Katsuya; Yanagisawa, Tadashi*; Narumi, Issei
no journal, ,
PprI, which is unique to 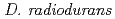 , is invoked by present data as the most important protein for radiation response mechanism. However, despite the interest in mechanism underlying the radiation response in
, is invoked by present data as the most important protein for radiation response mechanism. However, despite the interest in mechanism underlying the radiation response in 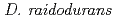 , little is known about the exact function of PprI protein. In this study, functional analysis of PprI protein was performed. Western blot analysis and gel shift assay suggested that PprI itself does not control directly the expression of
, little is known about the exact function of PprI protein. In this study, functional analysis of PprI protein was performed. Western blot analysis and gel shift assay suggested that PprI itself does not control directly the expression of 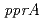 and
and 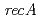 . We identified a novel regulatory protein PprM by 2D-PAGE.
. We identified a novel regulatory protein PprM by 2D-PAGE. 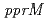 -disruptant strain showed significant sensitivity to
-disruptant strain showed significant sensitivity to  -rays. Western blot analysis revealed that PprM involves in RecA and PprA induction. By reporter assay it was found that PprM protein regulates the
-rays. Western blot analysis revealed that PprM involves in RecA and PprA induction. By reporter assay it was found that PprM protein regulates the 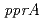 expression at the promoter level. These results suggest that PprM is involved in the unique radiation response mechanism controlled by PprI in
expression at the promoter level. These results suggest that PprM is involved in the unique radiation response mechanism controlled by PprI in 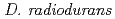 .
.
Sakashita, Tetsuya; Ikeda, Daisuke*; Hamada, Nobuyuki*; Suzuki, Michiyo*; Tsuji, Toshio*; Wada, Seiichi*; Funayama, Tomoo; Kobayashi, Yasuhiko
no journal, ,
Here we report how food-NaCl associative learning is affected by radiation exposure. Wild-type  strain was used for all experiments. The animals were irradiated with 0-500 Gy of
strain was used for all experiments. The animals were irradiated with 0-500 Gy of  -rays. Irradiation did not affect chemotaxis toward NaCl, indicating that sensing and signaling in chemo-attraction are maintained in irradiated animals. It has been reported that chemotaxis was decreased by the association of starvation with chemical stimulation of NaCl. Chemotaxis of animals irradiated during the food-starved conditioning with NaCl was decreased than that of non-irradiated control and temporarily stopped. At several hours after irradiation, the decrease of chemotaxis in irradiated animals was observed again. These results suggest that irradiation perturbed and temporarily stopped a part of neuron network related to associative learning, but not that to chemo-attraction to NaCl.
-rays. Irradiation did not affect chemotaxis toward NaCl, indicating that sensing and signaling in chemo-attraction are maintained in irradiated animals. It has been reported that chemotaxis was decreased by the association of starvation with chemical stimulation of NaCl. Chemotaxis of animals irradiated during the food-starved conditioning with NaCl was decreased than that of non-irradiated control and temporarily stopped. At several hours after irradiation, the decrease of chemotaxis in irradiated animals was observed again. These results suggest that irradiation perturbed and temporarily stopped a part of neuron network related to associative learning, but not that to chemo-attraction to NaCl.